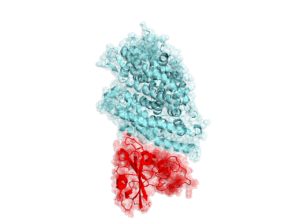
SARS-CoV-2 RBD domain in complex with human ACE2 receptor (PDBID: 6vsb, 6acg) [10.1126/science.abb2507, 10.1371/journal.ppat.1007236]
Using a more tradition top-down approach, IBM announced the formation of the COVID-19 High Performance Computing Consortium. Besides Big Blue, this includes basically all the big names you’d expect in the supercomputing space: The U.S. Department of Energy, the White House Office of Science and Technology Policy, MIT, Rensselaer Polytechnic Institute, Lawrence Livermore National Lab, Argonne National Lab, Oak Ridge National Laboratory, Sandia National Laboratory, Los Alamos National Laboratory, NASA, the National Science Foundation, Microsoft, Google, Hewlett Packard Enterprise, and Amazon.
Given the names on that list, it’s no surprise to see that the performance on tap is impressive. Across 16 sites, the consortium offers a combined 775,000 processor cores and 34,000 GPUs, good for 330 petaflops. Researchers can begin putting in proposals on how to use that power, with resources provided to researchers believed to “make the most immediate impact.”
On a more crowdsourced note, the Folding@Home project has been offering its distributing computer power to protein folding for medical research for years. Now the project is devoting its resources to COVID-19, and has seen a truly impressive response. In the last two weeks, they’ve seen 400,000 new contributors, a 1200% increase. All those new users have given the project a combined peak output of 470 petaflops.
The great thing is, medical research can benefit from both of these approaches, taking advantages of their unique strengths. The COVID-19 HPC Consortium can focus on directed research with an eye for immediate impact, while Folding@Home can fold those proteins with an eye for finding promising therapeutic compounds that are effective on COVID-19 protein. It’s easy to see these approaches at odds in other aspects, but it’s great seeing both methodologies at work for this important research.