Genomics research has become an indispensable part of biological research which can help diagnose multiple illnesses and determine individual therapies, or better identify viruses and fight more efficiently against them, as seen during the SARS-CoV-2 pandemic.
According to a fantastic research piece dating from 2015 and quoted on the National Human Genome Research Institute, it is estimated that genomics research will generate between 2 and 40 exabytes of data per year. Not only these numbers are staggering, but the historical growth rates are equally impressive: over the last decade, the total amount of sequence data produced doubled every seven months.
As genomics become the new normal in condition analysis and treatment, data requirements will become the elephant in the room. How can life sciences organizations address these data growth rates, and what challenges need to be addressed?
Data Processing in Genomics
From an IT perspective, genomics presents a unique convergence of challenges. First, data sources are heterogeneous: DNA sequencers are produced by several manufacturers, and each sequencer comes with its own suite of proprietary software tools, usually incompatible with those from the competitors.
Second, data sources are distributed: genome sequencing instruments are spread out at various locations in different countries; some are located in universities, others in hospitals, or in private research facilities. While not a challenge for private research labs doing their own sequencing, this can easily become a major pain point for research initiatives collecting data from scattered sequencing locations across multiple storage types, especially when massive datasets are involved. This makes it difficult to collect data in one single central place.
Data Transformation and Locality
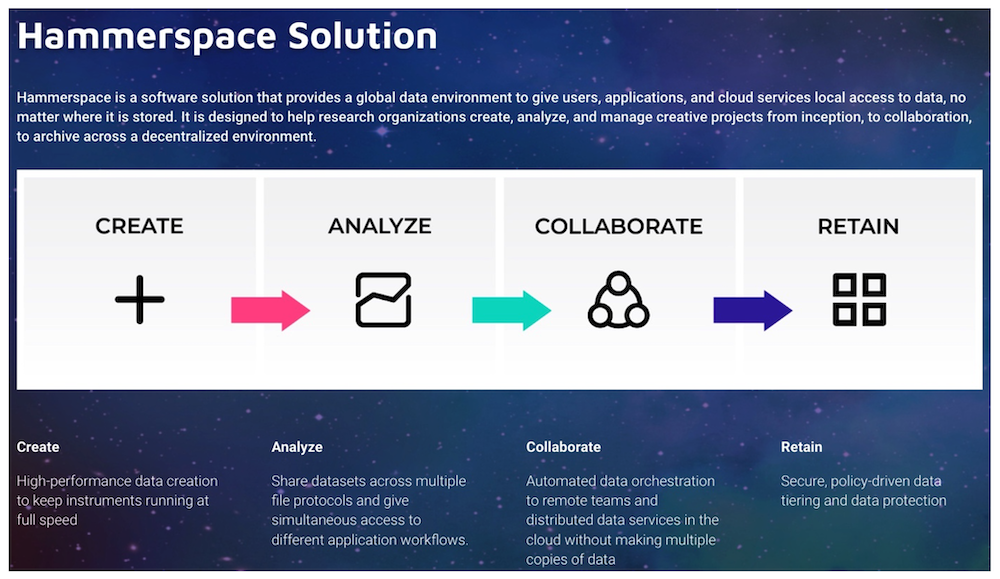
Yet another aspect of genomics is the number of steps required to obtain usable data. Raw data needs to be assembled, then is put through several transformation and normalization steps which may take hours, even days.
Sequencers output raw data, consisting of series of short DNA sequences, to expensive high performance storage. Data needs to be assembled and error-corrected via a process called multiplexing, which transforms raw data into readable, processed files. Once this is completed, data needs to be again verified for consistency validation, which also takes time. Finally, once validations are completed, data is ready to be provided to other applications to perform medical or scientifical research. This often requires data to move to other more affordable storage types, and/or to facilitate collaboration.
Each solution uses its own software, and some researchers may also want to use other proprietary or open-source solutions, often via one or more file protocols. The problem is that researchers need immediate global multi-protocol file access to the data, which by necessity might be stored on multiple storage systems. This increases operational complexity, which adds cost and inhibits collaboration.
Finally, data locality isn’t only about where the data is stored, but also to whom (the researchers) and to what (the various applications) it is accessible. Increasingly this often involves remote access across multiple sites and the Cloud. Without a global namespace that bridges incompatible data storage silos and locations that provides users with a continuous data plane, manageability of the datasets becomes extremely complex. Data gets siloed, and operational complexity is increased with multiple directories and mountpoints. This is made worse with the typical practice of copying data from one location to another to provide online access to the files for users and applications.
Characteristics of Storage Platforms for Genomics
To sum it up, the distributed and heterogeneous nature of genomics workloads drives up demand for a global distributed data platform capable of proposing the following features:
- A single global namespace capable of globally federating data stored across locations and incompatible storage types drastically simplifies data management and provides simple data access paths for users and applications alike, enabling global access and collaboration use cases.
- Data efficiency, advanced replication, and tiering mechanisms to avoid unnecessary and time-consuming data copies, ensure data is adequately protected, stored efficiently and on the right storage tier from a $/GB perspective.
- Multiprotocol file and object access methods to meet varied protocol requirements of DNA sequencers, demultiplexing applications, and other tools, as well as research and business applications that will leverage the data.
- Uncompromising performance to accelerate data processing and achieve faster time-to-value
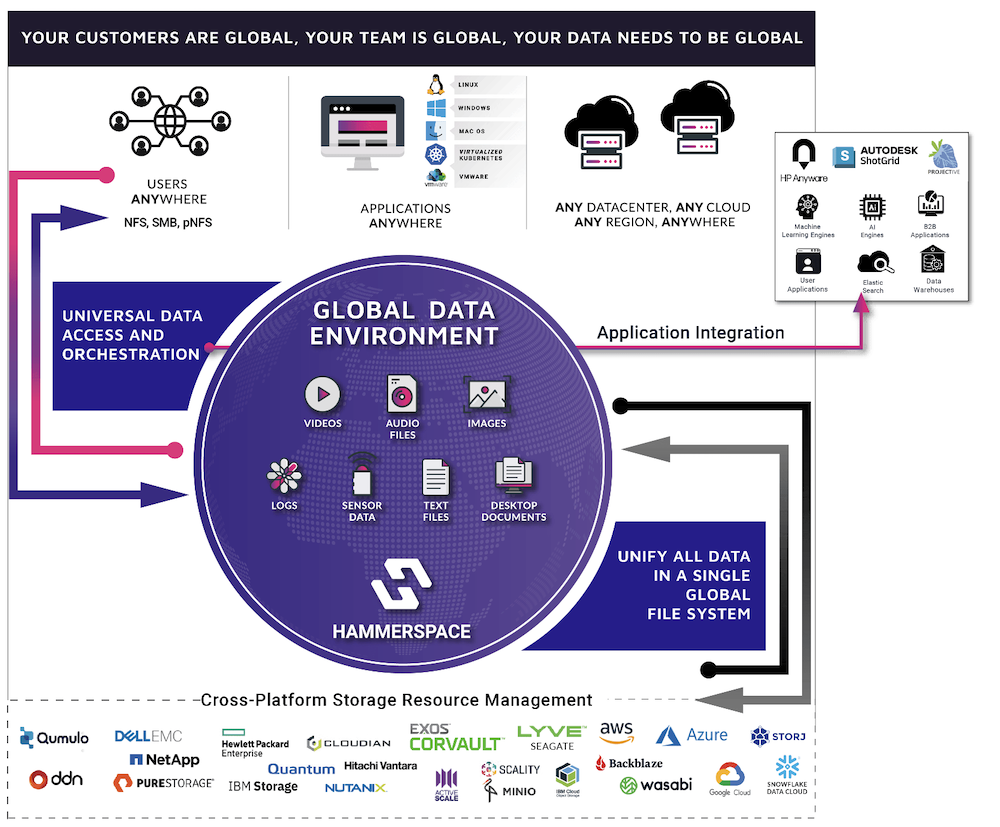
Hammerspace Global Data Environment is perfectly tailored to meet all these requirements, by offering a software-defined global filesystem that spans across any data center, any cloud, and any region and can aggregate the existing data on any legacy storage system into a single, shared global namespace.
The solution supports a broad spectrum of storage types from any vendor, enabling high-speed ingest rates and low latency on fast media for DNA sequencer output, as well as fast reads for demultiplexing and checksum tasks, reducing the length of processing jobs. Multi-vendor storage type flexibility also provides the ability to transparently tier data between high performance storage to capacity-oriented storage or cloud from any vendor, while still being available via the same global namespace, without disrupting user or application workflows.
The ability to provide online access to all data everywhere without the need to manage copies also eliminates wait times that can result from solutions that require long-running copy jobs from one location to another. This powerful capability further accelerates the execution of sequences in the data processing workflow.
Hammerspace also delivers a rich set of data services including anti-virus protection, undelete and file versioning, snapshots and clones, compression, deduplication, tiering and immutable file shares. It also embeds multiple security features including encryption, external KMS server support, role-based access control and Active Directory support.
Conclusion
Genomics is a highly specialized discipline which provides tremendous value to humanity, by providing insights and personalized treatments into medical conditions for humans and animals, and by being a precious ally to research in many other fields such as agriculture and virology, to name a few.
Due to its high value but also to its complexity, genomics data and workflows require a storage platform capable of meeting multiple challenges. An ideal solution should deliver performance and affordable capacity, a distributed data platform, and capabilities that eliminate low-value, time consuming data operations while accelerating data processing steps.
Hammerspace can deliver across all these requirements and more, thanks to a fast, distributed multiprotocol data platform which reduces complexity and operational costs in distributed and multi-vendor data environments. This helps both increase time to results for research pipelines, while also helping keep storage costs under control.